Creatures from the Black Lagoon: Lessons in the Diversity and Evolution of Eukaryotes
by Scott DawsonI’ve organized this lecture into several parts, beginning with a history of biological classification of life — especially in the context of microorganisms. I also want to talk about the diversity and lifestyle of microbes (protists in particular) that live without oxygen and why this is relevant to the evolution of life — especially with respect to eukaryotic evolution. Then I’m going to discuss how the majority of microbial life is yet to be discovered, and how one might find new microbes of interest to the study of evolution. And lastly, I’ll talk about some new local monsters we've found — microbial creatures from the Black Lagoon, where they live (just in case you want to check them out), and why they could change our understanding of eukaryotic evolution.
So, to start, I think when I first I learned about evolution there was always an implied "march of progress," or a linear progression from one form of life to a next, higher and higher, up the evolutionary ladder. This notion usually begins with some primordial soup — progressing to bacteria — to protist — to plant to animal — bugs — to us. But we know this is false. Evolution is branching — or at least that’s been the thinking in the past 150 years. I’ll be talking more about this, but in order to do good research in evolution, we need to examine where these evolutionary "stories" come from — what are the assumptions? I’m going to try to address several of these assumptions and misconceptions by showcasing some of the scientists who formed the basis of the classification of microorganisms.
Aristotle
Let's start in ancient Greece with Aristotle — in addition to many other
contributions to science, he was the first to articulate a dichotomy
of life, classifying all life as being either plants or animals. This
ancient world view of biology persists today, and misinforms our
perspective in terms of evolution, not to mention the structuring of
scientific research (like botany departments and zoology
departments). It was a few thousand years later that the microscope was
invented, and animal/vegetable/or mineral types of classification
were hardly enough to classifiy the new microorganisms, first
called "animacules." Microbes are often non-descript rods, or
spheres. How then does one classify microbes, particularly when they
don't really look different from one another like plants look
different from animals? More about how to do that later, but on to
more history.

|
|
Carolus Linnaeus Linnaeus lived in the 18th Century and created the familiar hierarchical classification scheme of life: kingdom, family, class, order, family, genus and species. Again, this classification scheme is based on how organisms looked — essentially their phenotype. Although this heirarchy within the plant and animal kingdoms was an improvement — an attempt to standardize classification whose legacy persists today — there were still no good working definitions of "kingdoms" or "species" which could apply to microorganisms when they were discovered. Even today the conception of a species of bacteria is hard to define, in the same way an animal species is defined. |

|
|
Charles Darwin We are all probably familiar with Charles Darwin, 19th century father of natural selection and evolution, but he also called for a framework or geneology of organisms in order to study evolution. Darwin helped to develop the concept of a geneology of life with branching lineages. But we attribute the conception of evolutionary relationships among organisms with a tree — a geneology — to Ernst Haeckel at roughly the same time. |
 |
|
Ernst Haeckel Ernst Haeckel also worked in the mid-19th century. Haeckel was the first to coin the term "protist" or "Protista," although his definition also included the bacteria. |
|
Haeckel's Tree of Life Ernst Haeckel is attributed to be the first to describe the evolutionary relationships among living organisms, a geneology of life, as analogous to a tree. Haeckel, interestingly, described all living things as falling into not just two kingdoms (plants and animals), but also a third kingdom for microorgansims — the Protista. There are several other interesting features of this vision — for example, Haeckel postulated a common origin for all life (plants, animals and microbes). This is still a common assumption, but it makes sense with modern molecular evidence as well. OK, let's fast forward a hundred years or so. How do we classify microbes today, especially when they have no particular morphology or shape to speak of. Scientists had all but given up on classifying microbes based on their evolutionary relationships (a phylogeny) until Carl Woese. |
|
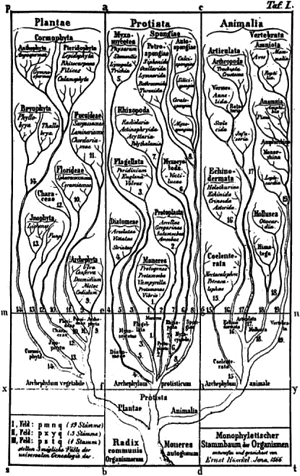
|
The Woesian Revolution Woese based his classification on molecules,
not how organisms look or act. This transition from classification
based on phenotype (taxonomy) to one based on genotype enabled him to
determine the evolutionary relationships (a phylogeny) among bacteria — something other researchers had all but given up on. Woese's work was founded on the principle suggested in 1965 that "molecules are
documents of evolutionary history." Basically, DNA can be thought of
as molecular fossils. At the University of Illinios in the late 1970s, Woese wanted to
determine evolutionary relationships among microorganisms, and in
the process, he and colleagues discovered a huge split in the
"prokaryotes" — as big a genetic difference as that between prokaryotes
and eukaryotes. Woese orignally thought that these were primitive
organisms and so called them the Archaea. Later studies showed that
the Archaea were actually more related to the eukaryotes in many
aspects that to the bacteria. So, rather than five kingdoms of life,
Woese argued for three domains (Eucarya, Archaea, and Bacteria). This
conception of three domains of life and two domains of prokaryotes, rather
than the standard prokaryote/eukaryote dichotomy, really shook up the
study of the evolution of life. So how did Woese arise at this
remarkable conclusion? First he needed a molecule to study from all
organisms — ribosomal RNA (rRNA).
Woese based his classification on molecules,
not how organisms look or act. This transition from classification
based on phenotype (taxonomy) to one based on genotype enabled him to
determine the evolutionary relationships (a phylogeny) among bacteria — something other researchers had all but given up on. Woese's work was founded on the principle suggested in 1965 that "molecules are
documents of evolutionary history." Basically, DNA can be thought of
as molecular fossils. At the University of Illinios in the late 1970s, Woese wanted to
determine evolutionary relationships among microorganisms, and in
the process, he and colleagues discovered a huge split in the
"prokaryotes" — as big a genetic difference as that between prokaryotes
and eukaryotes. Woese orignally thought that these were primitive
organisms and so called them the Archaea. Later studies showed that
the Archaea were actually more related to the eukaryotes in many
aspects that to the bacteria. So, rather than five kingdoms of life,
Woese argued for three domains (Eucarya, Archaea, and Bacteria). This
conception of three domains of life and two domains of prokaryotes, rather
than the standard prokaryote/eukaryote dichotomy, really shook up the
study of the evolution of life. So how did Woese arise at this
remarkable conclusion? First he needed a molecule to study from all
organisms — ribosomal RNA (rRNA).
|
||
|
Why use rRNA? Ribosomal RNA (rRNA) is an RNA component of the ribosome, the cellular machine which translates the DNA genetic code to amino acids, and subsequently, proteins. The rRNA genes are ubiquitous in all life, being conserved enough to identify, yet containing enough variabilty to determine evolutionary relationships. But Woese has had a hard time selling this idea, mostly because everyone has been trained to view life as prokaryotes/eukaryotes, plants/animals, or the updated version of that &emdash; the five kingdom view. |
|
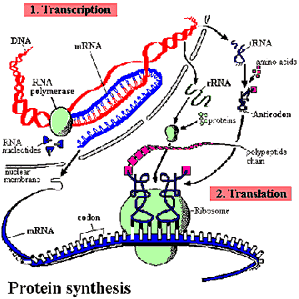 |
|
Five Kingdoms or Three Domains? In the 1960s, Whittaker added one kingdom (fungi) to Aristotles's view of life (plants and animals) but still relegated all microbes to either the Protista (eukaryotic microbes) or the Monera (bacteria). Something from this figure (and view) should be obvious. Why should we emphasize macroscopic life over microscopic life? In terms of life on Earth, we still live in an age of microbes. For the majority of geologic time, some 3-4 billion years, life has evolved — and at least three billion has been solely microbial. |
|
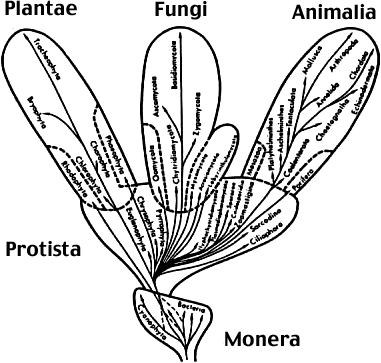 |
|
The five-kingdom view is still taught, as
I'm sure many of you know. Carl Woese started a revolution in the
classification of life, much like Copernicus did with his idea that the
Sun doesn't revolve around the Earth. We, as humans, possess a
lot of unique characteristics such as the ability to understand
the world we live in. But we are not the pinnacle of evolution — nothing is. Evolution is about change through time, not about
ranking life based on some criterion. Besides, since the bulk of
genetic diversity of life on Earth is been microbial (note where we
are in Woese's tree), shouldn't they get attention simply for that
reason? If you take nothing else from my talk, stop thinking about
biology in terms of five kingdoms. |
||
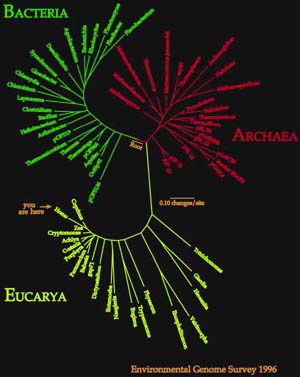
|
|
Since Woese's early trees, we've added a lot of sequences as you can see
in the figure to the left. I'd like to switch now from talking about
microbes in general, to specfically eukaryotic microbes (that is,
microbial cells which possess a membrane-bound nucleus and tend to be
unicellular). What's the current picture of eukaryotic
phylogeny?
|
|
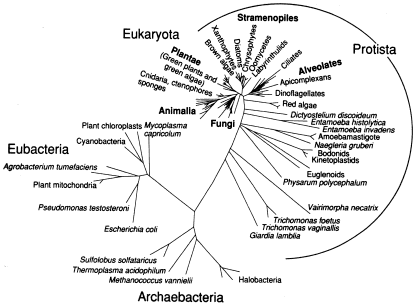
The Evolution of the Eukaryotes As in the prokaryotic world, one can see from this schematic tree of the eukaryotes (below), that the majority of eukaryotic life is, in fact, microbial (protists). There is far more phenotypic and genotypic diversity in all the protist groups combined than within the plant, animal or fungal kingdoms. How many kingdoms of eukaryotes are there? This is still largely unknown. If we use the genetic measure, we come up with roughly 70 eukaryotic kingdoms, only some of which are shown here. Further, because of our bias toward macroscopic life, most of the biology of these protists is largely unknown. How did the eukaryotes evolve? To understand that, we need to visualize the early Earth.
II. Life in anoxic worlds
Some of the oldest fossils belong to what look like photosynthetic bacteria — bacteria which breathe carbon dioxide and exhale oxygen. So this process happened early in Earth's history. Microbes made the world. Think about it — oxygen is actually quite poisonous, generating hydrogen peroxides and oxygen radicals. Many microbes don't have a way of dealing with poisonous oxygen. So they avoided it . . . and they still do today.
Contemporary
Virtually anywhere! Check it out — the
Black sea is the largest land-locked anoxic basin. Can we find more
organisms like Giardia which might still live in anoxic worlds, and
still be unknown to us? How could we do it?
|
|||||||
 A tablespoon of mud
A tablespoon of mudWe start with about a tablespoon of mud — remember that this can contain several billion microbes. This is mud from Berkeley Aquatic Park. Then we extract the DNA from the mud. By the way, the black color of the mud is a good indicator that there is no oxygen at that layer. Reduced iron complexed with sulfur produces something called pyrite — a dark black precipitate. |
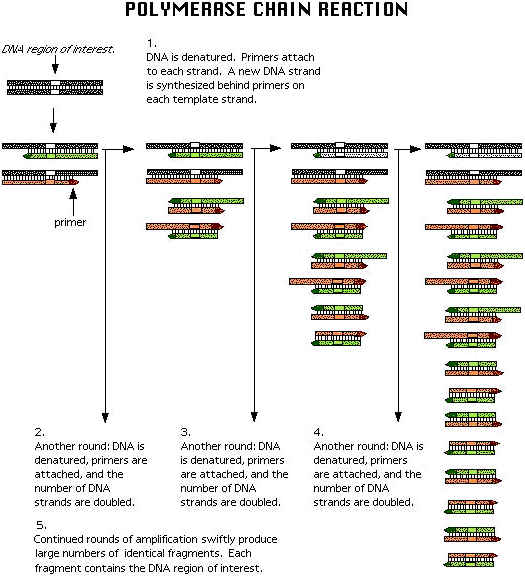
Xeroxing DNA
The polymerase chain reaction, or PCR, is
basically a way to xerox DNA so that we have enough to work with in
the lab. Many microbes are rare and we need to amplify their DNA
signals to be able to find them. In our work we're xeroxing a
specific gene — the gene for rRNA. Remember this is what Carl Woese
used to relate different microbes to each other and construct a tree
of life.
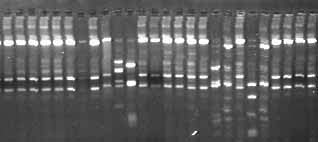 Patterns in DNA
Patterns in DNAOnce we amplify rRNA genes from a given environmental sample, we need to sort them by type. One tablespoon of mud could have several billion microbes, perhaps representing 1,000 different types! To categorize these different types, we use a technique called RFLP (restriction fragment length polymorphism) which generates unique patterns or fingerprints corresponding to unique organisms. At right is an example showing the DNA fingerprints of the different types of sequences (organisms) we find. Notice the different banding patterns.
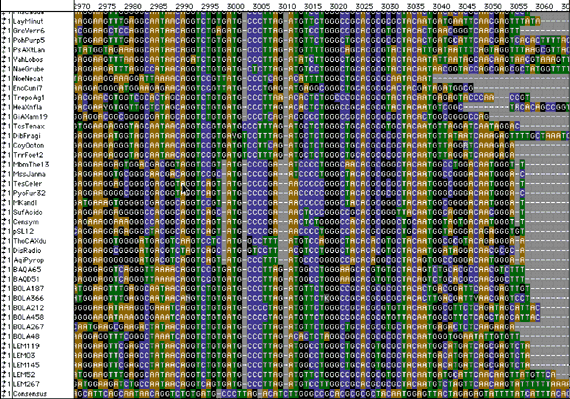
Aligning genes
Once we get the DNA sequence, we line up the
sequences and compare them to known organisms. The rows correspond to an
rRNA sequence from a particular organism; the column relates the exact
nucleotide (A,T,C,G) at a specific position in the sequence of the
gene. Note the conserved/variable regions.
Constructing a phylogenetic tree
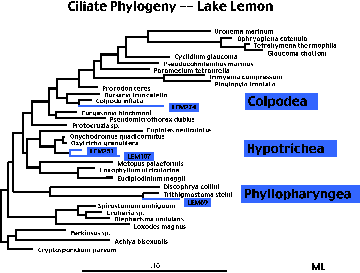 Then,
from this alignment of DNA rRNA sequences, we use computer models to
simulate the evolution of sequences. We infer that the evolution of
the sequences corresponds to the evolution of the organisms. In this
way, we are able to determine evolutionary relationships among the
new uncultivated microbes. Unlike Linneaus, Darwin or Haeckel, we
organize geneologies based on genotype, not phenotype. This also
gives us a picture of evolution. Note the organisms in blue — those
we only know from environmental sequences or fingerprints. Also note
how we can compare where they fall on the tree to known organisms. OK, let's talk more about our study of creatures from the Black Lagoon.
Then,
from this alignment of DNA rRNA sequences, we use computer models to
simulate the evolution of sequences. We infer that the evolution of
the sequences corresponds to the evolution of the organisms. In this
way, we are able to determine evolutionary relationships among the
new uncultivated microbes. Unlike Linneaus, Darwin or Haeckel, we
organize geneologies based on genotype, not phenotype. This also
gives us a picture of evolution. Note the organisms in blue — those
we only know from environmental sequences or fingerprints. Also note
how we can compare where they fall on the tree to known organisms. OK, let's talk more about our study of creatures from the Black Lagoon.
IV. Evolutionary Implications of Uncultivated Eukaryotes |
|
|
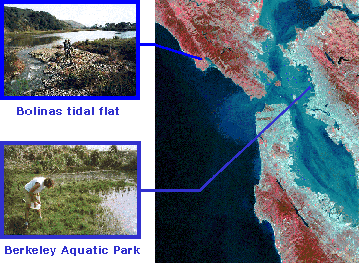
Marine Sampling Sites So we've chosen three sites altogether in which to check for previously undiscovered microbes. Two of the sites are local: Bolinas Lagoon by Stinson Beach and the Berkeley Aquatic Park. We chose these two sites because the sediment had low oxygen levels.
Freshwater Sampling |
|
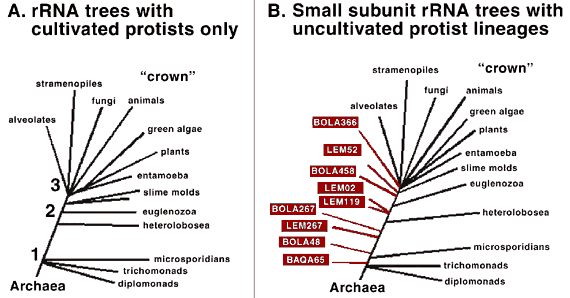
 Outside the "Crown"
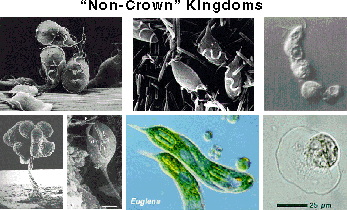
Outside the "Crown"Many of the protists we've found are not closely related to any known group of protists. The "crown" refers to the most recent "big bang" in eukaryote evolution — where it seems a lot of groups of organisms evolved quickly, including our animal ancestors. Several of the new environmental sequences are deep in the tree, suggesting that they might have retained some characteristics of the ancestral eukaryotes. We think we've found about eight new kingdoms of eukaryotes in just three environments by just hunting for their DNA. Not bad for a teaspoon of stinky mud. These pictures are of Giardia (top left), Trichomonas (top middle), a slime mold (bottom left), Euglena (bottom middle) and several amitochondriate amoebae (top and bottom right).
New Kingdoms of Eukaryotes
Models and Sampling Bias
Conclusions So, it's off to find new microbes — where should we look next?
|
|
 Early Earth
Early Earth Fossil Redbeds
Fossil Redbeds
 Cyanobacteria
Cyanobacteria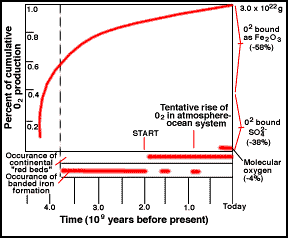 Evolution of Oxygen
Evolution of Oxygen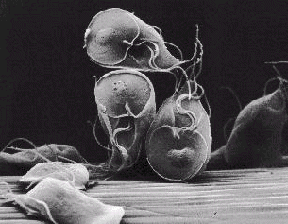 Giardia is an aquatic protist, and a parasite that major cause of dysentery
world wide. Hopefully no one here has encountered it while camping.
Giardia has no mitochondrion and it likes to avoid oxygen. Why?
Oxygen is an abundant source of energy, but in many aquatic
environments, it gets quickly used up by microorganisms. Many
bacteria, however, can respire or "breathe" alternatives to oxygen
— sulfur, hydrogen, nitrogen for example. These bacteria are
called anaerobes (more on this later). Environments lacking oxygen
(anoxic) harbor many of these types of bacteria &emdash; billions per
teaspoon of sediment or muck. In many cases, the anoxic environments
harbor more microorgansisms than oxic ones. Giardia survives by
eating anaerobic bacteria in these rich anoxic environments. But
where on Earth are these "extreme" anoxic environments? Where would
one go to find them?
Giardia is an aquatic protist, and a parasite that major cause of dysentery
world wide. Hopefully no one here has encountered it while camping.
Giardia has no mitochondrion and it likes to avoid oxygen. Why?
Oxygen is an abundant source of energy, but in many aquatic
environments, it gets quickly used up by microorganisms. Many
bacteria, however, can respire or "breathe" alternatives to oxygen
— sulfur, hydrogen, nitrogen for example. These bacteria are
called anaerobes (more on this later). Environments lacking oxygen
(anoxic) harbor many of these types of bacteria &emdash; billions per
teaspoon of sediment or muck. In many cases, the anoxic environments
harbor more microorgansisms than oxic ones. Giardia survives by
eating anaerobic bacteria in these rich anoxic environments. But
where on Earth are these "extreme" anoxic environments? Where would
one go to find them?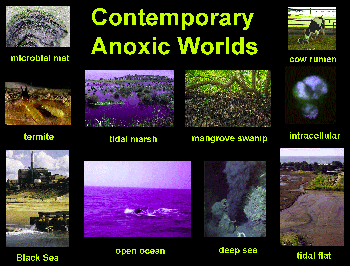
 Characteristics of eukaryotes that live without oxygen
Characteristics of eukaryotes that live without oxygen Identifying protists by microscopy
Identifying protists by microscopy Identifying protists by cultivation
Identifying protists by cultivation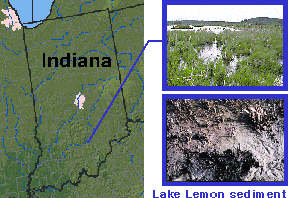
 Boiling Hot Springs!
Boiling Hot Springs!